Processing and integrating 5k PBMCs CITE-seq data
Contents
Processing and integrating 5k PBMCs CITE-seq data#
This notebooks provides an example for CITE-seq data processing in Python.
CITE-seq is a method for cellular indexing of transcriptomes and epitopes by sequencing. CITE-seq data is single-cell data comprising transcriptome-wide measurements for each cell (gene expression) as well as surface protein level information, typically for a few dozens of proteins. The method is described in Stoeckius et al., 2017 and also on the cite-seq.com website.
The data used in this notebook is on peripheral blood mononuclear cells (PBMCs) and has been provided by 10x Genomics.
[1]:
# Change directory to the root folder of the repository
import os
os.chdir("../")
Download data#
The data is available here. Both filtered and raw matrices are required for this notebook.
[2]:
# This is the directory where those files are downloaded to
data_dir = "data/pbmc5k_protein"
[3]:
for file in os.listdir(data_dir):
print(file)
filtered_feature_bc_matrix
raw_feature_bc_matrix
5k_pbmc_protein_v3_raw_feature_bc_matrix.tar.gz
5k_pbmc_protein_v3_filtered_feature_bc_matrix.tar.gz
Load libraries and data#
Import libraries:
[4]:
import numpy as np
import pandas as pd
import scanpy as sc
[5]:
import muon as mu
from muon import prot as pt
First, we will load filtered matrices that contain counts for cell but not empty droplets. These cells will be used for the downstream analysis.
[6]:
mdata = mu.read_10x_mtx(os.path.join(data_dir, "filtered_feature_bc_matrix"))
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1094: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if not is_categorical(df_full[k]):
Next, we will also load count matrices for all the droplets, whether containing cells or empty.
[7]:
mdata_raw = mu.read_10x_mtx(os.path.join(data_dir, "raw_feature_bc_matrix"))
Protein#
[8]:
prot = mdata.mod['prot']
prot
[8]:
AnnData object with n_obs × n_vars = 5247 × 32
var: 'gene_ids', 'feature_types'
DSB normalisation#
This normalisation method developed for CITE-seq data uses background droplets defined by low RNA content in order to estimate background protein signal and remove it from the data. More details and its original implementation are available in the ``dsb` GitHub repository <https://github.com/niaid/dsb>`__.
muon brings dsb normalisation method to Python CITE-seq workflows.
pt.pp.dsb(mdata, raw=mdata_raw, empty_droplets=droplets)
Please note due to the nature of the dsb method it behaves in a slightly different way than many other preprocessing and normalisation methods. Its implementation in muon offers 2 ways to use the method.
Operate on 2 AnnData / MuData objects — filtered and raw. Cell calling from the filtered data is used to define cells in the raw data then.
As in the original implementation, operate on raw matrices and then return a copy of the data where only specified cells are kept.
Here, we use the first way of using the method. We would still need to calculate log10umi value for each droplet to define which droplets are empty.
[9]:
mdata_raw['rna'].obs["log10umi"] = np.array(np.log10(mdata_raw['rna'].X.sum(axis=1) + 1)).reshape(-1)
[10]:
mu.pl.histogram(mdata_raw['rna'], ['log10umi'], bins=50)
/usr/local/lib/python3.8/site-packages/seaborn/distributions.py:369: UserWarning: Default bandwidth for data is 0; skipping density estimation.
warnings.warn(msg, UserWarning)
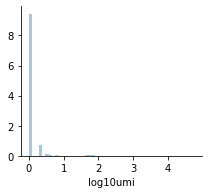
For clarity, let’s zoom into the part of this data:
[11]:
mu.pl.histogram(mdata_raw['rna'][mdata_raw['rna'].obs.log10umi >= 1], ['log10umi'], bins=50)
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1094: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if not is_categorical(df_full[k]):
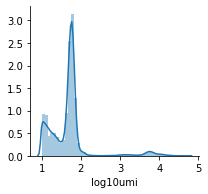
In this dataset, isotype antibody controls are available that also can be taken into account by the normalisation method:
[12]:
isotypes = mdata_raw['prot'].var_names[29:32].values
isotypes
[12]:
array(['IgG1_control_TotalSeqB', 'IgG2a_control_TotalSeqB',
'IgG2b_control_TotalSeqB'], dtype=object)
Preserve original counts in a layer before the normalisation:
[13]:
prot.layers['counts'] = prot.X
Normalise counts in mdata:
[14]:
pt.pp.dsb(mdata, mdata_raw, empty_counts_range=(1.5, 2.8), isotype_controls=isotypes, random_state=1)
/Users/bredikhi/git/compbio/muon/muon/_prot/preproc.py:129: UserWarning: Dropping 49 empty droplets as they are already defined as cells
warn(
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1094: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if not is_categorical(df_full[k]):
Plot values to visualise the effect of normalisation:
[15]:
sc.pl.scatter(mdata['prot'], x="CD3_TotalSeqB", y="CD19_TotalSeqB", layers='counts')
sc.pl.scatter(mdata['prot'], x="CD3_TotalSeqB", y="CD19_TotalSeqB")
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
... storing 'feature_types' as categorical
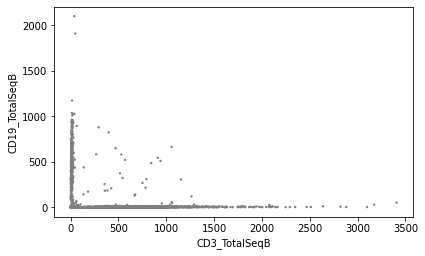
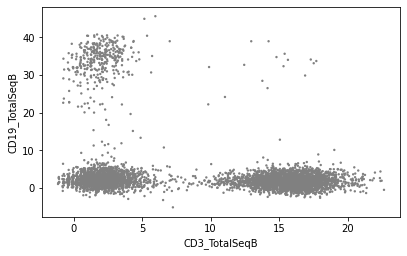
Downstream analysis#
We can run conventional methods like PCA on the normalised protein counts:
[18]:
sc.tl.pca(prot)
[19]:
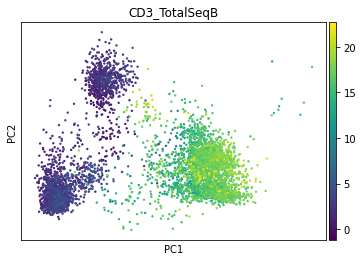
sc.pl.pca(prot, color='CD3_TotalSeqB')
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
[20]:
sc.pp.neighbors(prot)
sc.tl.umap(prot, random_state=1)
[21]:
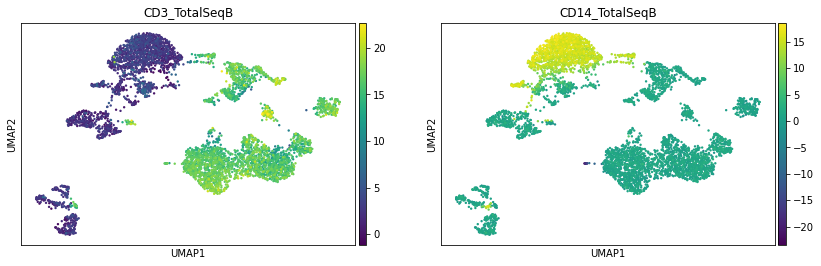
sc.pl.umap(prot, color=['CD3_TotalSeqB', 'CD14_TotalSeqB'])
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
RNA#
[22]:
rna = mdata.mod['rna']
rna
[22]:
AnnData object with n_obs × n_vars = 5247 × 33538
var: 'gene_ids', 'feature_types'
QC#
Perform some quality control. For now, we will filter out cells that do not pass QC.
[23]:
rna.var['mt'] = rna.var_names.str.startswith('MT-') # annotate the group of mitochondrial genes as 'mt'
sc.pp.calculate_qc_metrics(rna, qc_vars=['mt'], percent_top=None, log1p=False, inplace=True)
[24]:
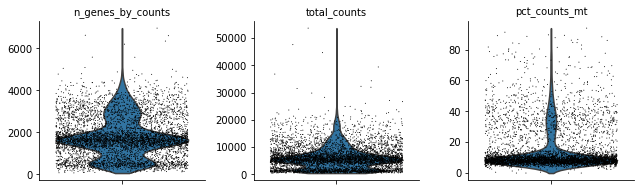
sc.pl.violin(rna, ['n_genes_by_counts', 'total_counts', 'pct_counts_mt'],
jitter=0.4, multi_panel=True)
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
... storing 'feature_types' as categorical
Filter genes which expression is not detected:
[25]:
mu.pp.filter_var(rna, 'n_cells_by_counts', lambda x: x >= 3)
# Same as the following but doesn't copy the object:
# sc.pp.filter_genes(rna, min_cells=3)
Filter cells:
[26]:
mu.pp.filter_obs(rna, 'n_genes_by_counts', lambda x: (x >= 200) & (x < 5000))
# Same as the following but doesn't copy the object
# sc.pp.filter_cells(rna, min_genes=200)
# rna = rna[rna.obs.n_genes_by_counts < 5000, :]
mu.pp.filter_obs(rna, 'total_counts', lambda x: (x > 1500) & (x < 15000))
mu.pp.filter_obs(rna, 'pct_counts_mt', lambda x: x < 20)
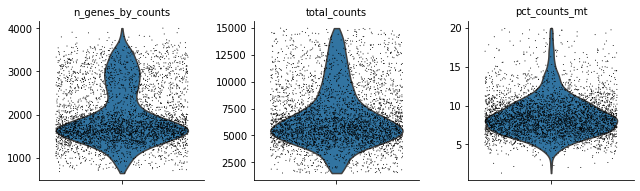
Let’s see how the data looks after filtering:
[27]:
sc.pl.violin(rna, ['n_genes_by_counts', 'total_counts', 'pct_counts_mt'],
jitter=0.4, multi_panel=True)
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
Normalisation#
We’ll normalise the data so that we get log-normalised counts to work with.
[28]:
sc.pp.normalize_total(rna, target_sum=1e4)
[29]:
sc.pp.log1p(rna)
Feature selection#
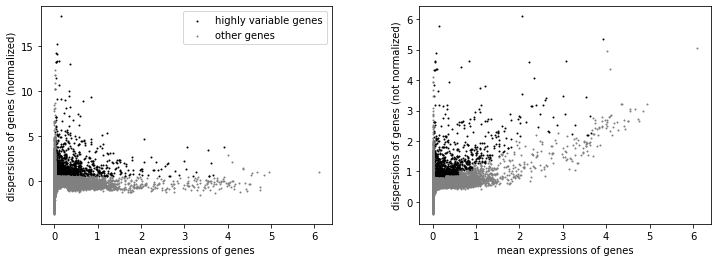
We will label highly variable genes that we’ll use for downstream analysis.
[30]:
sc.pp.highly_variable_genes(rna, min_mean=0.02, max_mean=4, min_disp=0.5)
[31]:
sc.pl.highly_variable_genes(rna)
[32]:
np.sum(rna.var.highly_variable)
[32]:
1734
Scaling#
We’ll save log-normalised counts in a .raw slot:
[33]:
rna.raw = rna
… and scale the log-normalised counts to zero mean and unit variance:
[34]:
sc.pp.scale(rna, max_value=10)
Analysis#
Having filtered low-quality cells, normalised the counts matrix, and performed feature selection, we can already use this data for multimodal integration.
However it is usually a good idea to study individual modalities as well. Below we run PCA on the scaled matrix, compute cell neighbourhood graph, and perform clustering to define cell types.
PCA and neighbourhood graph#
[35]:
sc.tl.pca(rna, svd_solver='arpack')
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1094: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if not is_categorical(df_full[k]):
To visualise the result, we will use some markers for (large-scale) cell populations we expect to see such as T cells and NK cells (CD2), B cells (CD79A), and KLF4 (monocytes).
[36]:
sc.pl.pca(rna, color=['CD2', 'CD79A', 'KLF4', 'IRF8'])
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
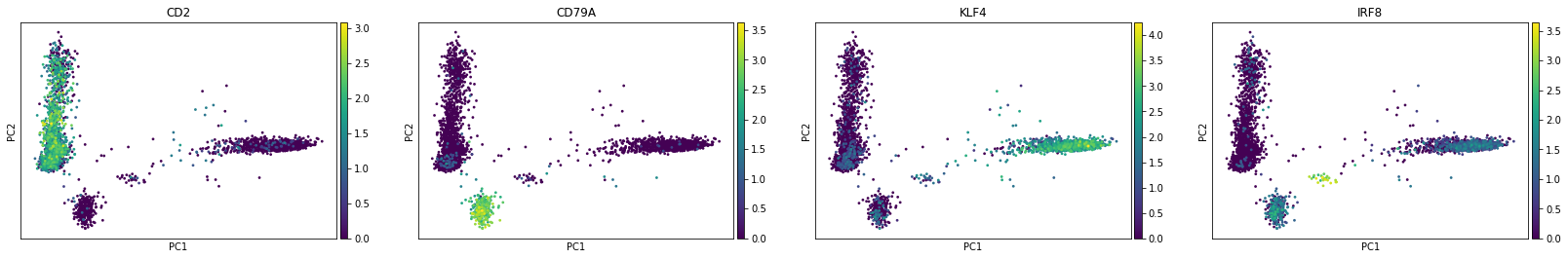
The first principal component (PC1) is separating myeloid (monocytes) and lymphoid (T, B, NK) cells while B cells-related features seem to drive the second one. Also we see plasmocytoid dendritic cells (marked by IRF8) being close to B cells along the PC2.
[37]:
sc.pl.pca_variance_ratio(rna, log=True)
Now we can compute a neighbourhood graph for cells:
[38]:
sc.pp.neighbors(rna, n_neighbors=10, n_pcs=20)
Non-linear dimensionality reduction and clustering#
With the neighbourhood graph computed, we can now perform clustering. We will use leiden clustering as an example.
[39]:
sc.tl.leiden(rna, resolution=.75)
To visualise the results, we’ll first generate a 2D latent space with cells that we can colour according to their cluster assignment.
[40]:
sc.tl.umap(rna, spread=1., min_dist=.5, random_state=11)
[41]:
sc.pl.umap(rna, color="leiden", legend_loc="on data")
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
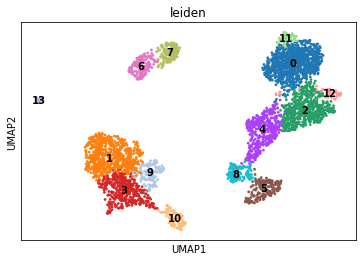
Cell type annotation#
[42]:
sc.tl.rank_genes_groups(rna, 'leiden', method='t-test_overestim_var')
[43]:
result = rna.uns['rank_genes_groups']
groups = result['names'].dtype.names
pd.set_option('display.max_columns', 50)
pd.DataFrame(
{group + '_' + key[:1]: result[key][group]
for group in groups for key in ['names', 'pvals']}).head(10)
[43]:
| 0_n | 0_p | 1_n | 1_p | 2_n | 2_p | 3_n | 3_p | 4_n | 4_p | 5_n | 5_p | 6_n | 6_p | 7_n | 7_p | 8_n | 8_p | 9_n | 9_p | 10_n | 10_p | 11_n | 11_p | 12_n | 12_p | 13_n | 13_p | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | RPL32 | 1.380112e-247 | S100A8 | 0.000000e+00 | IL7R | 4.827268e-175 | CST3 | 8.457789e-138 | CCL5 | 1.614990e-191 | GNLY | 1.868870e-151 | CD79A | 6.409675e-113 | IGHM | 1.284390e-142 | GNLY | 2.305955e-92 | TYMP | 4.113971e-44 | FCGR3A | 7.931312e-73 | CD8B | 1.344611e-28 | IL32 | 5.583487e-09 | ITM2C | 1.106098e-36 |
| 1 | RPS3A | 1.627373e-256 | S100A9 | 4.101797e-317 | LTB | 1.387967e-107 | CPVL | 4.945355e-162 | NKG7 | 2.032769e-140 | NKG7 | 5.122287e-96 | MS4A1 | 1.513867e-103 | CD79A | 6.570059e-124 | FGFBP2 | 1.190382e-102 | LYZ | 5.675880e-35 | LST1 | 3.169949e-44 | CD8A | 1.179723e-20 | TIGIT | 7.622707e-07 | CCDC50 | 3.365550e-34 |
| 2 | RPL30 | 9.687924e-237 | VCAN | 3.394343e-312 | IL32 | 2.100285e-104 | LYZ | 2.891312e-127 | GZMA | 1.524691e-106 | KLRD1 | 4.177880e-111 | HLA-DQA1 | 3.751459e-106 | CD79B | 2.500853e-106 | GZMH | 7.659337e-98 | FCN1 | 4.341500e-35 | TCF7L2 | 1.098160e-45 | RPS3A | 3.524939e-13 | PBXIP1 | 6.263715e-07 | IL3RA | 2.391788e-23 |
| 3 | RPL13 | 2.673413e-227 | LYZ | 1.467838e-237 | TRAC | 7.044805e-82 | FCN1 | 3.664234e-123 | CST7 | 2.410078e-106 | KLRF1 | 2.123803e-102 | BANK1 | 1.331708e-84 | IGHD | 9.719686e-72 | PRF1 | 1.626375e-91 | XAF1 | 7.801962e-43 | SMIM25 | 3.171766e-49 | LINC02446 | 7.812095e-13 | IKZF2 | 1.872429e-06 | PLD4 | 1.246058e-27 |
| 4 | RPL11 | 5.299188e-232 | MNDA | 1.131567e-278 | CD2 | 2.779068e-67 | FGL2 | 1.029085e-127 | IL32 | 2.124480e-90 | PRF1 | 1.035347e-103 | CD79B | 7.403563e-79 | MS4A1 | 5.945622e-89 | KLRC2 | 3.624602e-75 | FGL2 | 1.961404e-38 | LYN | 1.090891e-46 | RPS2 | 1.307254e-12 | TRBC2 | 1.364242e-06 | IRF8 | 2.272772e-29 |
| 5 | RPS12 | 2.484028e-223 | S100A12 | 3.040667e-307 | AQP3 | 1.363019e-56 | KLF4 | 5.745357e-131 | GZMM | 3.587211e-83 | CTSW | 2.070524e-100 | HLA-DQB1 | 6.762216e-69 | TCL1A | 5.585904e-65 | NKG7 | 5.237177e-70 | CST3 | 3.191349e-34 | MS4A7 | 6.730315e-51 | RPS6 | 1.514270e-12 | DUSP4 | 4.791963e-06 | TCF4 | 2.379937e-28 |
| 6 | RPS6 | 1.943468e-227 | FCN1 | 3.913932e-244 | CD3E | 4.321252e-53 | AIF1 | 3.486286e-117 | KLRG1 | 1.147997e-74 | GZMA | 8.167419e-91 | CD37 | 1.360556e-57 | HLA-DQA1 | 1.723135e-87 | CST7 | 5.730287e-82 | CTSS | 1.003140e-33 | CDKN1C | 2.347009e-39 | RPL13 | 1.869602e-12 | HACD1 | 1.189909e-05 | UGCG | 2.010952e-26 |
| 7 | RPL9 | 3.701542e-226 | CD14 | 5.684490e-266 | KLRB1 | 8.857507e-50 | HLA-DRA | 1.229331e-117 | CD3D | 3.637893e-73 | GZMB | 2.485553e-88 | RALGPS2 | 1.241454e-54 | CD37 | 6.685736e-66 | GZMB | 1.041564e-90 | MNDA | 1.065729e-35 | FCER1G | 4.274695e-38 | RPS12 | 2.277374e-12 | IL2RA | 1.689073e-05 | IRF7 | 3.697920e-27 |
| 8 | TPT1 | 4.096112e-227 | CTSS | 1.729932e-207 | RPS18 | 1.695589e-47 | PSAP | 1.832311e-117 | B2M | 7.472893e-70 | CLIC3 | 3.298338e-75 | HLA-DPB1 | 4.220414e-51 | HLA-DPB1 | 4.708672e-47 | CCL5 | 2.700983e-64 | PSAP | 1.983579e-33 | LRRC25 | 3.008410e-45 | RPL32 | 4.908655e-12 | FOXP3 | 2.616250e-05 | JCHAIN | 2.922103e-22 |
| 9 | RPL34 | 4.347625e-218 | CSF3R | 9.101933e-243 | SPOCK2 | 6.018993e-48 | HLA-DRB1 | 1.410033e-114 | GZMK | 1.345830e-53 | HOPX | 2.743326e-83 | HLA-DRA | 2.174338e-45 | CD74 | 2.262129e-43 | FCGR3A | 3.320130e-76 | CPVL | 3.518379e-38 | CFD | 5.047007e-41 | RPL28 | 4.009718e-12 | TRAC | 1.275863e-05 | MZB1 | 4.527073e-20 |
[44]:
new_cluster_names = {
"0": "CD4+ naïve T", "12": "Treg", "2": "CD4+ memory T",
"11": "CD8+ naïve T", "4": "CD8+ memory T",
"5": "NK", "8": "memory-like NK",
"6": "mature B", "7": "pre-B",
"1": "CD14 mono", "3": "intermediate mono", "9": "intermediate mono", "10": "CD16 mono",
"13": "pDC",
}
rna.obs['celltype'] = rna.obs.leiden.astype("str").values
rna.obs.celltype = rna.obs.celltype.replace(new_cluster_names)
rna.obs.celltype = rna.obs.celltype.astype("category")
We will also re-order categories for the next plots:
[45]:
rna.obs.celltype.cat.reorder_categories([
'CD4+ naïve T', 'CD4+ memory T', 'Treg',
'CD8+ naïve T', 'CD8+ memory T', 'NK', 'memory-like NK',
'pre-B', 'mature B',
'CD14 mono', 'intermediate mono', 'CD16 mono',
'pDC'], inplace=True)
mdata.update()
… and take colours from a palette:
[46]:
import matplotlib
import matplotlib.pyplot as plt
cmap = plt.get_cmap('rainbow')
colors = cmap(np.linspace(0, 1, len(rna.obs.celltype.cat.categories)))
rna.uns["celltype_colors"] = list(map(matplotlib.colors.to_hex, colors))
[47]:
sc.pl.umap(rna, color="celltype", legend_loc="on data", frameon=False)
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1192: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if is_string_dtype(df[key]) and not is_categorical(df[key])
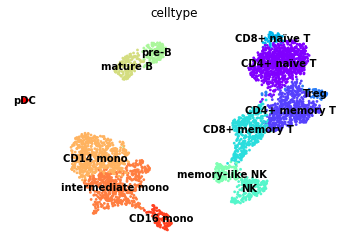
[48]:
sc.pl.umap(rna, color="celltype", legend_loc="on data", frameon=False,
title="RNA celltype annotation")
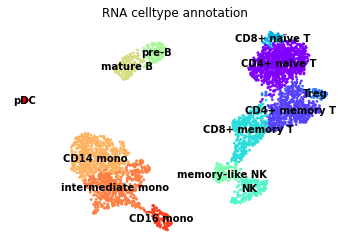
Finally, we’ll visualise some marker genes across cell types.
[49]:
marker_genes = ['IL7R', 'TRAC',
'ITGB1', # CD29
'FOXP3', 'IL2RA',
'CD8A', 'CD8B', 'CCL5',
'GNLY', 'NKG7', 'KLRC2',
'CD79A', 'MS4A1', 'TCL1A', 'IGHM', 'IGHD',
'IL4R', 'TCL1A',
'KLF4', 'LYZ', 'S100A8', 'ITGAM', # CD11b
'CD14', 'FCGR3A', 'MS4A7',
'CST3', 'IRF8', 'TCF4']
[50]:
sc.pl.dotplot(rna, marker_genes, groupby='celltype');
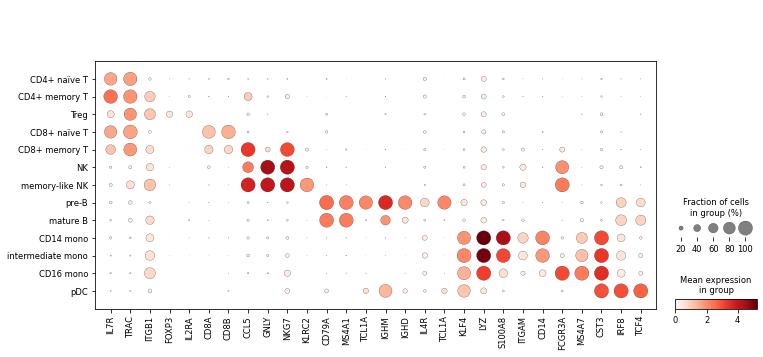
Plotting#
With mu.pl.embedding interface we can display an embedding from an individual modality (e.g. 'rna') and colour cells by a feature (variable) from another modality.
While variables names should be unique across all modalities, all individual modalities as well as the mdata object itself can have e.g. UMAP basis computed. To point to a basis from a specific modality, use mod:basis syntax, e.g. with "rna:X_umap" in the example below mdata.mod['rna'].obsm["X_umap"] basis is going to be used.
[51]:
mu.pl.embedding(mdata, basis="rna:X_umap", color=["CD3_TotalSeqB"])
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1094: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if not is_categorical(df_full[k]):

Multi-omics integration#
Having filtered cells at the RNA QC step, we will now subset them in the protein modality as well so that we can work with the union of cells:
[52]:
mu.pp.intersect_obs(mdata)
Multiplex clustering#
We can run clustering based on neighbours information from each modality taking advantage of multiplex versions of algorithms such as leiden or louvain.
[53]:
mu.tl.louvain(mdata, resolution=[2, .1], random_state=1)
[54]:
mu.tl.leiden(mdata, resolution=[2, .1], random_state=1)
[55]:
mdata.uns['louvain']
[55]:
{'params': {'resolution': [2, 0.1],
'random_state': 1,
'partition_improvement': 91323.3393340929}}
[56]:
mdata.uns['leiden']
[56]:
{'params': {'resolution': [2, 0.1],
'random_state': 1,
'partition_improvement': 91166.75577710925}}
[57]:
mu.pl.embedding(mdata, basis="rna:X_umap", color=["CD3_TotalSeqB", "louvain", "leiden"])
/usr/local/lib/python3.8/site-packages/anndata/_core/anndata.py:1094: FutureWarning: is_categorical is deprecated and will be removed in a future version. Use is_categorical_dtype instead
if not is_categorical(df_full[k]):
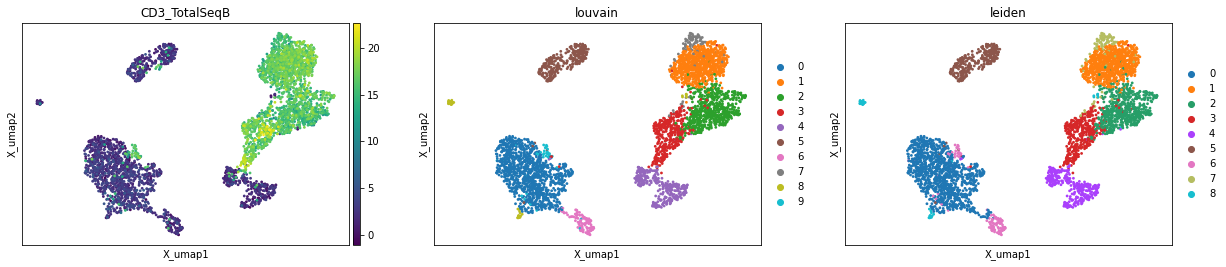
Multi-omics factor analysis#
To generate an interpretable latent space for both 'rna' and 'prot' modalities we will now run multi-omic factor analysis — a group factor analysis method that will allow us to learn an interpretable latent space jointly on both modalities. Intuitively, it can be viewed as a generalisation of PCA for multi-omics data. More information about this method can be found on the MOFA website.
[58]:
prot.var["highly_variable"] = True
mdata.update()
[21]:
mu.tl.mofa(mdata, outfile="models/pbmc5k_citeseq.hdf5",
n_factors=30)
#########################################################
### __ __ ____ ______ ###
### | \/ |/ __ \| ____/\ _ ###
### | \ / | | | | |__ / \ _| |_ ###
### | |\/| | | | | __/ /\ \_ _| ###
### | | | | |__| | | / ____ \|_| ###
### |_| |_|\____/|_|/_/ \_\ ###
### ###
#########################################################
Loaded view='prot' group='group1' with N=3891 samples and D=32 features...
Loaded view='rna' group='group1' with N=3891 samples and D=1734 features...
Model options:
- Automatic Relevance Determination prior on the factors: True
- Automatic Relevance Determination prior on the weights: True
- Spike-and-slab prior on the factors: False
- Spike-and-slab prior on the weights: True
Likelihoods:
- View 0 (prot): gaussian
- View 1 (rna): gaussian
######################################
## Training the model with seed 1 ##
######################################
Converged!
#######################
## Training finished ##
#######################
Warning: Output file models/pbmc5k_citeseq_v2.hdf5 already exists, it will be replaced
Saving model in models/pbmc5k_citeseq_v2.hdf5...
Saved MOFA embeddings in .obsm['X_mofa'] slot and their loadings in .varm['LFs'].
[62]:
# # NOTE: if you wish to load the trained model,
# # use mofax library to quickly add
# # factors and weights matrices
# # to the mdata object
# #
# import mofax as mfx
# model = mfx.mofa_model('models/pbmc5k_citeseq_v2.hdf5')
# mdata.obsm["X_mofa"] = model.get_factors()
# # If only highly variable features were used
# w = model.get_weights()
# # Set the weights of features that were not used to zero
# mdata.varm["LFs"] = np.zeros(shape=(mdata.n_vars, w.shape[1]))
# mdata.varm["LFs"][mdata.var["highly_variable"]] = w
# model.close()
[63]:
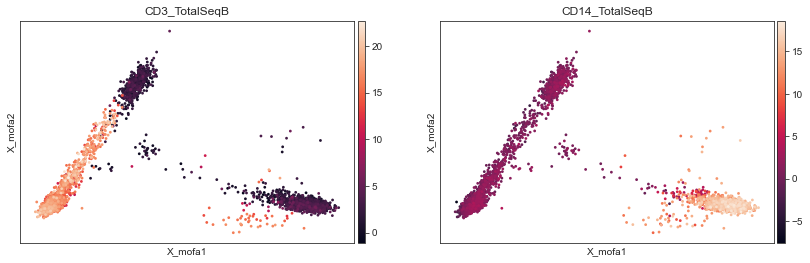
mu.pl.mofa(mdata, color=['CD3_TotalSeqB', 'CD14_TotalSeqB'])
... storing 'rna:leiden' as categorical
... storing 'rna:celltype' as categorical
... storing 'louvain' as categorical
... storing 'leiden' as categorical
[64]:
sc.pp.neighbors(mdata, use_rep="X_mofa")
sc.tl.umap(mdata, random_state=1)
[65]:
mdata.uns["rna:celltype_colors"] = list(map(matplotlib.colors.to_hex, colors))
[73]:
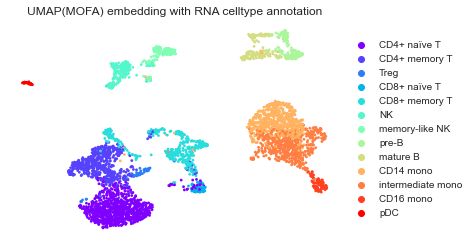
mu.pl.umap(mdata, color=['rna:celltype'], frameon=False,
title="UMAP(MOFA) embedding with RNA celltype annotation")
... storing 'louvain' as categorical
... storing 'leiden' as categorical
[74]:
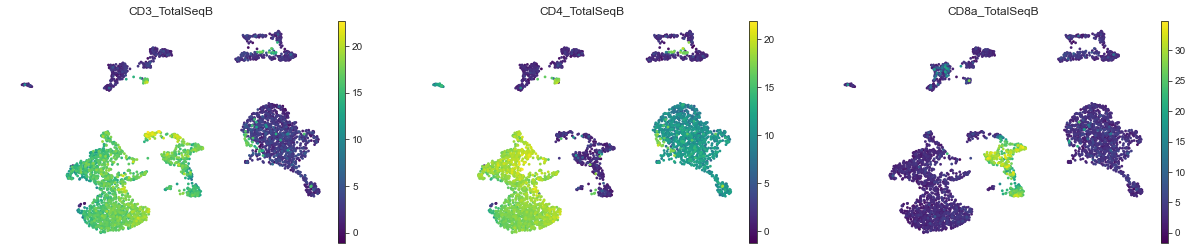
mu.pl.umap(mdata, color=['CD3_TotalSeqB', 'CD4_TotalSeqB', 'CD8a_TotalSeqB'], frameon=False, cmap='viridis')
... storing 'louvain' as categorical
... storing 'leiden' as categorical
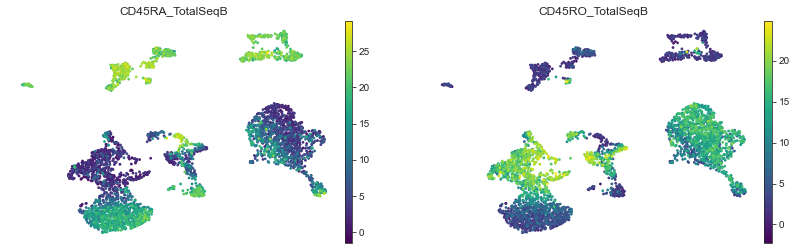
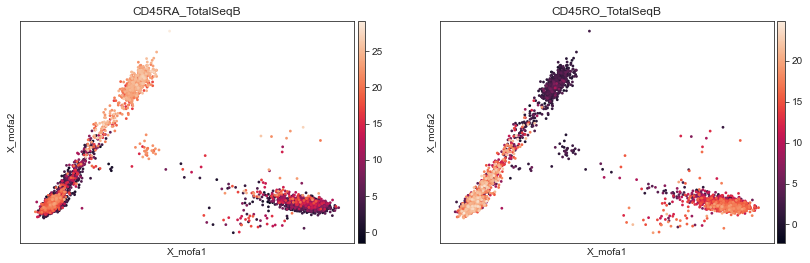
Proteins highlighting the memory — naïve axis will also help to refine memory/naïve T cells annotation:
[75]:
mu.pl.umap(mdata, color=['CD45RA_TotalSeqB', 'CD45RO_TotalSeqB'], frameon=False, cmap='viridis')
... storing 'louvain' as categorical
... storing 'leiden' as categorical
[76]:
mu.pl.mofa(mdata, color=['CD45RA_TotalSeqB', 'CD45RO_TotalSeqB'])
... storing 'louvain' as categorical
... storing 'leiden' as categorical
Interpreting the model#
[80]:
from matplotlib import rcParams
rcParams['figure.dpi'] = 150
[78]:
import mofax as mofa
model = mofa.mofa_model("models/pbmc5k_citeseq.hdf5")
[81]:
mofa.plot_weights(model, factors=range(3), n_features=10, sharex=False)
[81]:
<AxesSubplot:title={'center':'rna'}, xlabel='Feature weight'>
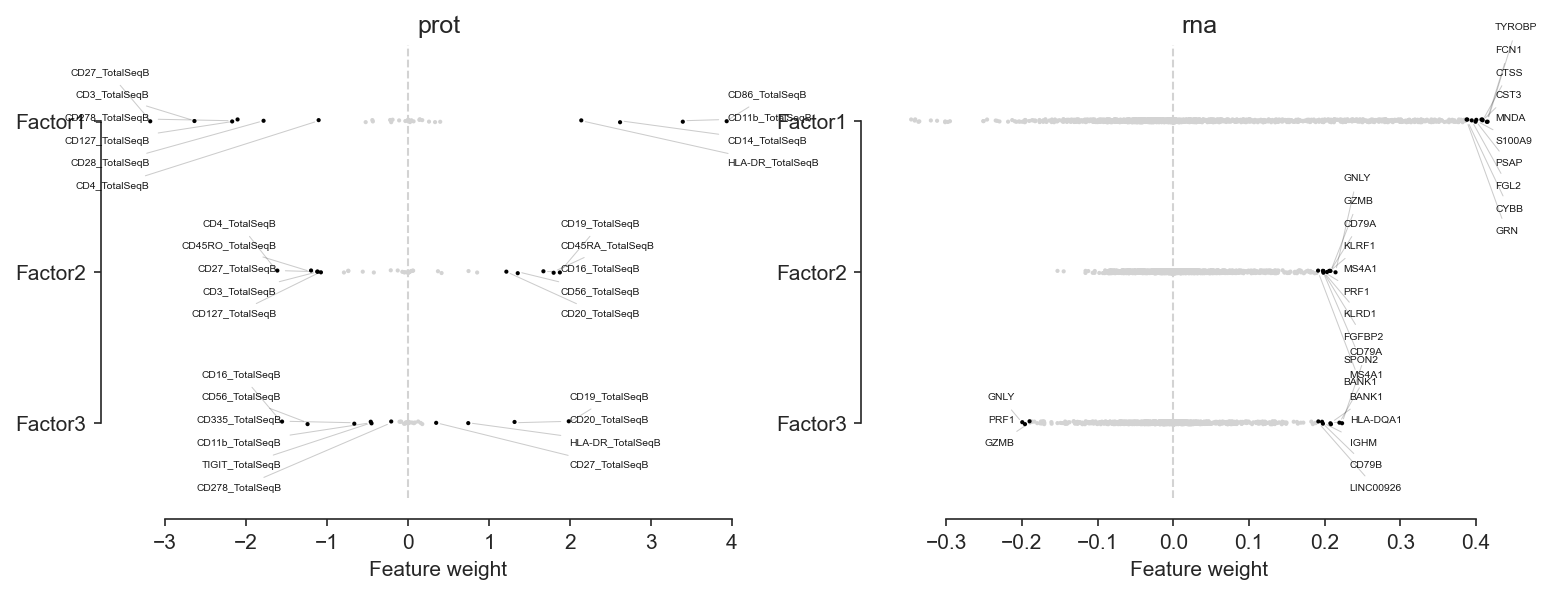
While Factor1 corresponds to lymphoid — myeloid axis, Factor2 seems to highlight T — B & NK axis and, at least partly, memory — naïve variability: CD45RO and CD127 are memory markers while CD45RA is a naïveté marker.
[84]:
model.metadata["rna:celltype"] = model.metadata["rna:celltype"].astype('category').cat.reorder_categories(mdata['rna'].obs.celltype.cat.categories)
[85]:
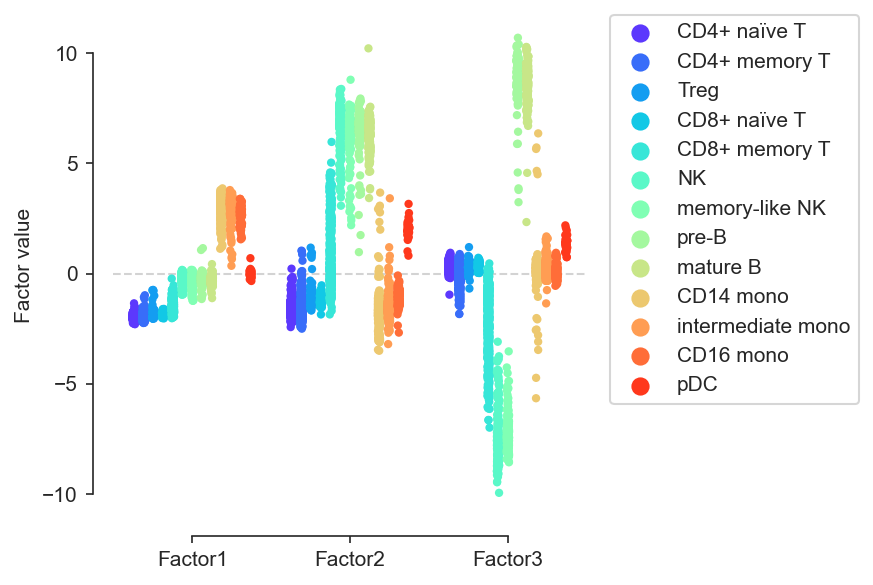
mofa.plot_factors_violin(model, color='rna:celltype', factors=range(3), dots=True, violins=False, palette='rainbow')
[85]:
<AxesSubplot:ylabel='Factor value'>
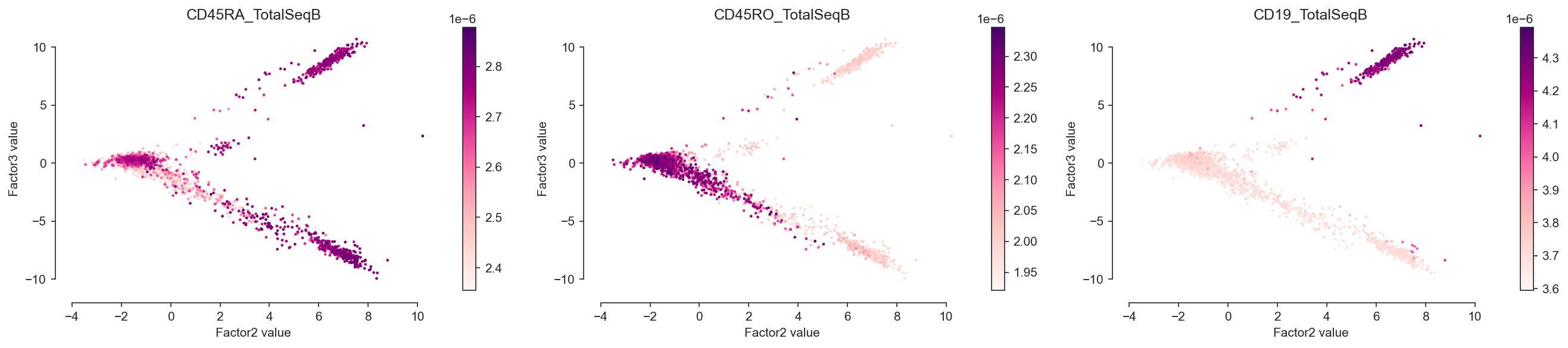
[90]:
mofa.plot_factors(model, x=1, y=2, color=["CD45RA_TotalSeqB", "CD45RO_TotalSeqB", "CD19_TotalSeqB"], palette="RdPu")
[90]:
<AxesSubplot:title={'center':'CD19_TotalSeqB'}, xlabel='Factor2 value', ylabel='Factor3 value'>
[91]:
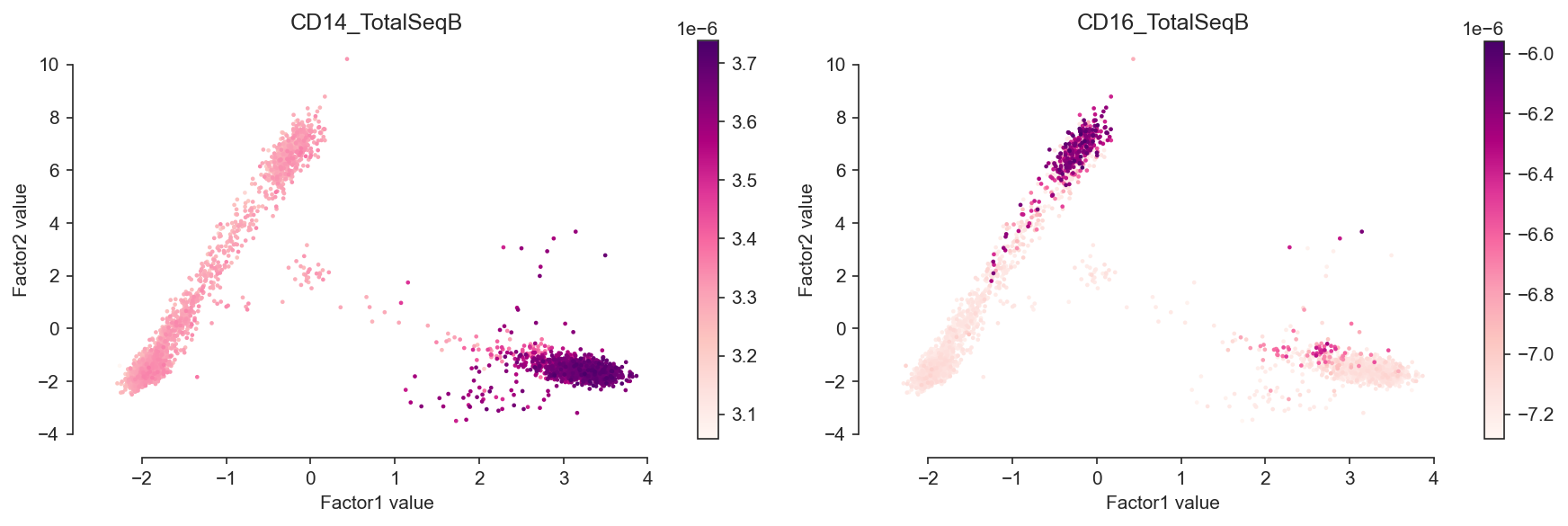
mfx.plot_factors(model, color=["CD14_TotalSeqB", "CD16_TotalSeqB"], palette="RdPu")
[91]:
<AxesSubplot:title={'center':'CD16_TotalSeqB'}, xlabel='Factor1 value', ylabel='Factor2 value'>
[95]:
model.close()
Saving multimodal data on disk#
We will now write mdata object to an .h5mu file.
[96]:
mdata.write("data/pbmc5k_citeseq.h5mu")
... storing 'louvain' as categorical
... storing 'leiden' as categorical
... storing 'feature_types' as categorical